http://www.youtube.com/watch?v=ok9esggzN18
ENZYMES
Enzymes are Biological catalysts (proteins) that speed up a chemical reaction by lowering its activity. Enzymes are not consumed during a reaction. One such example is catalase which is an enzyme that is found in all living cells. This enzyme breaks down hydrogen peroxide (H2O2).
Chemical reaction:
2H2O2 à 2H2O + O2.
The enzyme catalase breaks down H2O2 into water and oxygen.
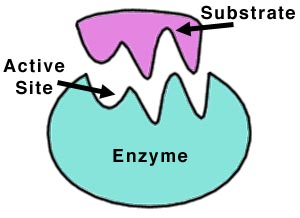
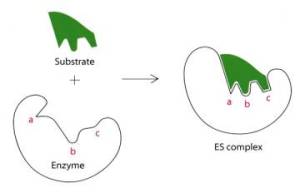
Ebzymes contains active site, where substrate fits into. This substrate will be catalysed. The substrate in this example above was H2O2.
Substrates bind to the active site of the enzyme by two theories;
- Lock and key – this is where the substrate is identically shaped to the active site and fits perfectly into the active site
- Induced fit – this is where the substrate is not identical to the active site but still binds to the enzyme for a reaction to occur.
Enzymes are turned on (activated) and turned off (deactivated or inhibited)
Enzymes are turned off/ inhibited;
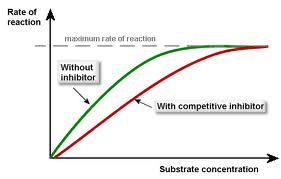
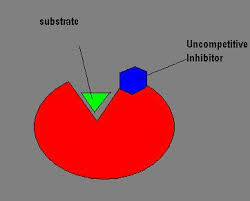
This is done by inhibition. It can either be competitive or non competitive inhibition. Competitive inhibition is when the inhibitor competes with the substrate for the active site of an enzyme and non competitive is where the inhibitor binds to another part (allosteric site)of the enzyme and causes a conformational change of the enzyme.
Enzymes are activated by;
- They are only produced when needed – this is a regulating method for reactions. These enzymes are activated when needed.
- Enzymes are activated using either cofactors (inorganic) or coenzymes (organic). These are small that are added to enzymes to activate the enzymes.
- Example of cofactor – Heme contained in hemoglobin
- Example of coenzyme – Thiamine (Vitamin B1)
Enzyme reactions are affected by;
- Substrate concentration
- Enzyme concentration
- Temperature
- pH
- Competitive inhibitors
- Non competitive/ allosteric inhibitors
Example of enzyme reaction vs temperature
As temperature increases, enzyme reaction increases until optimum temperature. Optimum temperature is the temperature or range of temperatures that the enzymes functions at its best. After this optimum temperature is passed enzymes becomes denatured and rate of reaction decreases as enzymes cannot bind to substrates due to conformational changes and breaking of chemical bonds within the active site and entire enzyme.